Infectious Diseases >> Application >> Influenza
Prepared in collaboration with Jason White
Outline of Sections
Section 1 |
|
Section 2 |
|
2.1 |
|
2.2 |
|
2.3 |
|
2.4 |
|
2.5 |
|
2.6 |
|
Section 3 |
|
3.1 |
|
3.2 |
|
3.3 |
|
3.4 |
|
3.5 |
|
3.5.1 |
|
3.5.2 |
|
3.5.3 |
|
3.5.4 |
|
3.6 |
|
3.7 |
|
Section 4 |
|
4.1 |
|
4.2 |
|
4.3 |
|
4.4 |
|
Section 5 |
|
5.1 |
|
5.2 |
|
5.3 |
|
Section 6 |
|
1 |
Introduction |
Influenza is a major cause of sickness and death around the world and is one of the most important infectious diseases confronting the world today. Combined with pneumonia, influenza is one of the ten leading causes of death in the United States. Even though most of its victims are elderly, pneumonia-influenza is one of the top-ten leading infectious conditions listed as causing years of potential life lost by the Centers for Disease Control. These facts, plus the memory of the great pandemic of 1918-1919, which killed about 550,000 people in the United States and claimed over 20 million lives around the world, have encouraged much research on this viral infection. The origins of virulent strains and the behavior of epidemics are still not well understood, however, and the unpredictability of emergence of new strains influenza proteins presents serious problems for public health-based control programs throughout the world.
One major complication to controlling the disease is the fact that flu, during the early phase of the infection, is often confused with the common cold. Influenza is a respiratory infection characterized by headaches, fevers, and short dry coughs. Usually, the disease lasts for up to a week, but infection is especially dangerous for the very young and the elderly, as well as those with certain chronic health problems. The elderly tend to have an overall lower resistance in general, and thus are more susceptible to influenza and its complications. Occasionally, there is a neuro-psychological complication known as “post-flu depression” that can last for several weeks or even months.
|
|
2 |
History |
Influenza was discovered not by a direct study of the disease in humans, but rather from studies on animal diseases. In 1918, J.S. Koen, a veterinarian, observed a disease in pigs which was believed to be the same disease as the now famous “Spanish” influenza pandemic of 1918.
In 1928, N. McBryde attempted to transmit influenza from pig to pig by taking mucus from the respiratory tract of sick pigs and instilling it into the noses of healthy pigs. However, this effort failed to transmit the disease because they inoculated only filtered material. Most types of bacterial filters used during that time were unable to allow the flow of viral particles through them.
During the same year, Robert Shope, working at the Rockefeller Institute for Comparative Pathology in Princeton, New Jersey, repeated the same experiments inoculating unfiltered material, instead. Shope was not only able to reproduce the disease, but more importantly, he went on to demonstrate that pigs can also become ill when filtered fluid was used for inoculation. The disease produced by the filtrate was mild and could be transferred repeatedly in pigs. This was the first reliable experimental evidence that influenza was caused by a virus, and it also provided the basis for further research into the human form of the disease.
2.1 |
Origins of Influenza |
People have suffered from influenza for thousands of years. Viruses identical or closely related to the human form of the infection can be isolated from ducks, turkeys, swine, horses, and many other species of warm-blooded vertebrates, as well. It is hypothesized that humans probably acquired influenza when domestication of animals first began. The development of agriculture and the establishment of permanent settlements provided sufficient numbers of potential hosts to create an epidemic. The term “influenza” was coined by an Italian in the mid-1700’s to conote a disease resulting from miasma (bad air). The human disease is thought to have arisen about 6000 years ago. A human influenza virus was not isolated until 1933. Wilson Smith, Christopher Andrews, and Patrick Laidrow first identified the virus that causes human influenza only because they found a suitable host for propagation. Laidrow used ferrets in his studies on canine distemper virus, and was able to propagate the flu virus in that same host, as well. Influenza virus was later shown to also infect adult mice and chicken embryos.
2.2 |
The Pandemic of 1580 |
The 1580 pandemic was the first documented outbreak in which we can be reasonably sure it was caused by the influenza virus. During the summer of 1580, influenza was reported in Asia Minor and Northern Africa. Italian accounts suggest that it spread from Malta to Sicily in July 1580 and subsequently diffused north through the Italian peninsula by August. During that time, Phillip II ruled the Iberian Peninsula, Southern Italy, and several North African ports. The early July outbreak in the Spanish Netherlands was most likely caused by troops sent by Phillip II to fight the Dutch.
2.3 |
Influenza Pandemics During the Eighteenth Century |
The medical profession of the eighteenth century was ill equipped to deal with influenza at any level. For most doctors, especially in the first six or seven decades of the century, influenza seemed to be spread or "created" by atmospheric factors. Such theories were complex. They usually invoked an unknown poison carried in the air and/or specific winds, temperature changes, barometric pressures, or other meteorological factors to explain the appearance and spread of endemic diseases like influenza. To that extent, most eighteenth and early nineteenth-century articles on influenza are much more likely to contain elaborate meteorological tables than geographical reconstructions or statistics on morbidity or mortality. Influenza pandemics occurred at least three times in the eighteenth century: 1729-1730, 1732-1733, and 1781-1782. In addition there were two major epidemics that could possibly be considered pandemics, in 1761-1762 and 1788-1789. Of these, two warrent further discussion: the pandemic of 1729-1730 and the great pandemic of 1781-1782.
The 1729-1730 pandemic was the first recorded pandemic, most likely fostered by the age of discovery. Flu apparently did not break out in North America until October 1732, when the disease was widespread along the New England coast from Boston to southern Maine. Although the origins and termination of the 1729-1730 outbreak are unclear, it obviously was a pandemic, the first of a series that western European observers perceived as coming from Russia. An origin in Russia seems plausible, but there is no documentation of this. Initial reports were of substantial outbreaks in two widely separated Russian cities, Moscow and Astrakhan, on the Caspian Sea, in April, 1729. There were no further reports during the summer, but influenza prevailed in Sweden in September and in Vienna in October. During November, flu was prevalent in Hungary and Poland, swept deep into Germany and appeared in London, Plymouth, York and Durham England, as well in Dublin, Ireland.
While quantitative evidence is lacking, the 1729-1730 pandemic caused sickness but relatively few deaths. Morbidity was extensive, but mortality was generally low, although the case-fatality rate was considered serious in northern Italy. Persons of all ages were stricken, but deaths were most numerous among the elderly and pregnant women.
The pandemic of 1781-1782 ranks with those of 1889-1890 and 1918-1919 as amongst the most widespread and dramatic outbreaks of disease in history. Unlike other pandemics of the eighteenth century, the pandemic of 1781-1782 had some interesting features that can be compared to the epidemics of the 20th century. A few general characteristics of this pandemic were noticed.
- evidence of a spring wave in North Africa and North America in 1781.
- diffusion of influenza into the Eastern Hemisphere in 1781.
- widespread outbreaks occurred within China and British-occupied India during the autumn of 1781.
- the pandemic started in China and spread westward in 1782.
It caused tens of millions of cases, spread as rapidly as existing transportation systems permitted, and not surprisingly, elicited volumes of medical writings.
2.4 |
Influenza Pandemic of 1918 |
The influenza pandemic of 1918, also known as “Spanish” flu, killed more rapidly than any other form of influenza known up to that time. This particular strain of influenza was especially dangerous to young adults. The National Office of Vital Statistics in the United States recorded the distribution of deaths during 1917 showing that the highest levels of morbidity occurred at the extremes of infancy and old age, and was very low in between these two age groups. By 1918, morbidity changed dramatically, and was now highest for the very young, and higher yet for persons between the ages of twenty and forty. The elderly appeared to be spared, for the most part. Thus, the Spanish Flu had two outstanding characteristics: 1) it killed millions of people and 2) most of them were in the prime of their life.
This distinctive strain of influenza swept across the face of the earth in three major waves between 1918 and 1919. Although it is uncertain as to where the first wave in the spring of 1918 originated, all available evidence indicates that it appeared in the United States in March of 1918. It attracted very little attention because pneumonic complications were few and deaths even fewer; it appeared as no more than just another bout with the kind of respiratory disease that so often circulates during that time of the year. Only later, after the second and third killer waves appeared did statisticians notice that an unusually large proportion of the flu and pneumonia victims were young adults.
The second wave occurred during March and April and expanded across North America, temporarily disrupting the operation of some military camps and a few factories. It was during this wave that the disease spread throughout most of the rest of the world. According to records, the disease reached epidemic proportions in Europe in April. It swept across the continent in the spring and summer, and the number of casualties was devastating. In Switzerland alone during the month of July, 53,000 people succumbed to the Spanish Flu.
In late August, the severity of the infection changed, suddenly transforming into the most dangerous strain (or strains) ever recorded. It occurred in three major parts of the North Atlantic almost simultaneously: Freetown. Sierra Leone, where local West Africans were brought together with British, South African, East African, and Australian soldiers and sailors; Brest, France, which was the chief port for Allied troops; and Boston, Massachusetts, one of America’s busiest embarkation ports and a major crossroads for military and civilian personnel of every nation involved with the Allied war effort. Massive troop movements and the disruption of significant segments of the population during World War I played an important role in the transmission of the disease.
2.5 |
The Origins of the 1957 H2N2 Pandemic |
In May 1957, an epidemic of a disease presumed to be influenza was reported in Hong Kong. After several months of investigation, most epidemiologists agreed that a strain of influenza virus had surfaced in China early in 1957. Meanwhile, the disease was spreading outward from Southeast Asia.
The new strain of influenza, initially referred to as “Asian Flu” had rapidly spread from Hong Kong to Japan, the Philippines, Malaya, and Indonesia by the end of May 1957. By June, there were numerous reports of influenza among passengers and crew on board ships that had departed from East Asian ports. During June, the disease also spread though India and the Middle East. Port cities were among the first places to be affected. Given the nature of international trade, the disease rapidly spread towards England, and reached the United States.
The Asian strain of influenza continued to be the most pronounced type A variant internationally for more than a decade. It was at this time that epidemiologists and biostatisticians seriously started to explore methods of devising an early warning detection system, at least within the context of determining the severity of an influenza epidemic and its geographical locale.
2.6 |
The 1976 Swine Flu "Epidemic" |
The details surrounding the swine-influenza "outbreak" of 1976 is notable because the event lead to a major change in public health policy. While the exact origins and validity of the Swine Flu outbreak are unknown, many public health and medical researchers believed that the type of swine influenza that was recovered from human victims during the January 1976 outbreak at Fort Dix, New Jersey was so closely related to the influenza strain that caused more than 500,000 deaths in the United States during the 1918-19 pandemic that to ignore the possibility of a repeat of that epidemic seemed irresponsible.
As the result, a nationwide inoculation program was instituted. However, the establishment of such a program was not easy to implement. In spite of identification of swine flu the proposal for a nationwide inoculation program gained little support at the federal level during April and May of that year. Although President Gerald Ford espoused the need for such a program, there was a consensus among many Congressmen that such a program was not warranted. The vaccine program did not officially begin until October. As history would show, the predicted epidemic failed to materialize. However, millions became vaccinated, some with devastaing health consequences. Guillian-Barre Syndrom, a decending paralysis, was not one of the predicted outcomes, yet occurred with a high degree of frequency at the height of the vaccination program. The vaccine was produced by Merck, Inc., but the insurance policy agaist such an eventuality was underwritten by the United States govenment. The public ended up paying out millions of dollars in health care benefits to affected individuals.
3 |
Biology Of The Influenza Virus |
3.1 |
Virulence Factors |
Influenza has presented humankind with a moving target since its appearance on the planet, but remakably, the clinical aspects have remained fairly constant. A given human influenza virus can evolve rapidly by one of two mechanisms: 1) antigenic shift (genetic reassortment between a human and a non-human virus in a non-human host), and 2) antigenic drift (accumulation of mutations that facilitate evasion of the host immune response). New influenza viruses constantly emerge from the environment, emanating from such disparate sources as migratory waterfowl, swine, domestic poultry, and sea mammals.
Reassortment of viral genomes within these animals is the genetic equivalent to recombination in eukaryotic cells, and is an important mechanism of antigenic variation. A pair of viral genes code for two surface glycoproteins, haemagglutinin (H) and neuraminidase (N), the crucial antigens against which the host develops immunological defenses. Thirteen H and nine N subtypes have so far been discovered, many of which occur only in various species of birds, particularly waterfowl. Types H1, H2, H3, N1, and N2 have been positively linked to epidemics in man.
Between major epidemics, mutations produce minor but cumulative antigenic variants in a wide number of animal reservoirs. Major new viral subtypes have appeared in 1918, 1957, and 1968, in each case resulting in a pandemic. The term “pandemic” literally means a worldwide epidemic, but for influenza there is also the assumption that a major subtype involving new H and/or N antigens has appeared. These shifts are much more dramatic than the normal slow genetic drift of other subtypes. As a result there is no opportunity for people to develop immunity to the new virus thus allowing the virus to spread faster.
Potentially pandemic strains probably arise from recombination with viruses from animal reservoirs. A wide range of virus subtypes have been discovered in wild and domesticated animals, most notably in pigs, horses, turkeys, and ducks. Feral and domesticated ducks harbor an enormous array of antigenic types and may play a major role in the ecology of the influenza A virus. In this century there have been four major shifts: 1918 (Hsw1N1=H1N1, 1957 (H2N2), 1968 (H3N2) and a very puzzling reappearance of a 1950 strain of H1N1, which began to circulate in 1977. There is some evidence that only a limited number of antigenic types recirculate over time in human populations.
Influenza viruses are unique in their ability to cause both recurrent annual epidemics and more serious pandemics that spread rapidly and may affect all or most age groups. The size of epidemics and pandemics are a direct result of antigenic variation of the virus, the amount of protective immunity in populations, and the relative virulence of the viruses.
3.2 |
Molecular Biology |
All organisms whose cells contain a nucleus in their cells, together with many bacteria and viruses, base their reproduction on a DNA (deoxyribonucleic acid) genome. However, the influenza virus is among a minority of organisms that base their replication on RNA. The properties of RNA help to explain the epidemiology of influenza and the difficulty of achieving successful prophylactic treatment by using vaccines.
DNA and RNA genomes both use polymerase enzymes for replication, but under suitable conditions, RNA molecules can replicate spontaneously and maintain continuous synthesis. Between the two types of genome, DNA and RNA, RNA viruses have the ability to evolve a million times more rapidly than their DNA based host. Because the error rate of RNA is so high, many variants of an RNA virus may coexist and compete. The fittest of these variants and hence the best adapted to its host will become the most abundant within a given population.
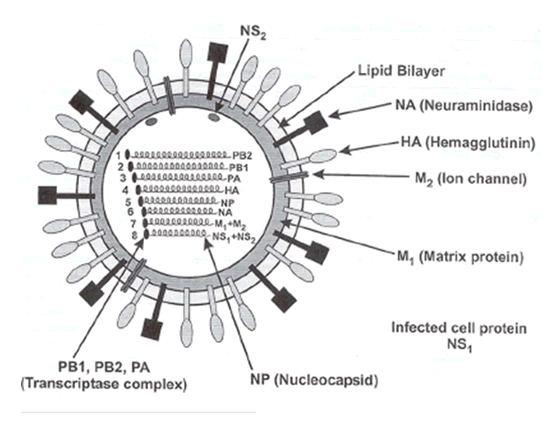
Influenza is caused by an orthomyxovirus, measuring 80 to 120 nanometers in diameter. The core of the viral particle consists of ribonucleoproteins surrounded by a lipid envelope that contains two types of glycoproteins in the form of spikes. The haemagglutinin spikes are responsible for binding the virus to red blood cells and other host cells, as well. The second protein, neuraminidase is an enzyme that cleaves terminal sialic acid residues from host-membrane bound glycoproteins and glycolipids. The neuraminidase facilitates release of viral progeny from infected cells as well as the spread of the virus from one cell to another. While antibodies against both haemagglutinin and neuraminidase may reduce the reproduction of the virus in the host, only the antibodies against the haemagglutinin can neutralize the infectivity of the virus particle. There are three strains of flu virus, A, B, and C, each characterized by the antigenic properties of its internal, nonglycosylated components.
3.3 |
A Strain Viruses |
Type A influenza causes pandemics, while influenza viral types B and C are less important in causing human disease, mainly infecting animals. Unfortunately, influenza A strains are the most varied of the three types. They have been isolated from mammalian, avian, and human sources. The influenza A genome consists of eight separate pieces of single-stranded RNA. These eight segments function as distinct genes, so genetic reassortment occurs readily when a host cell is infected with two strains of the virus. Influenza B and C strains have been isolated from human infections. The B strains of the virus are less lethal and often cause illness within more youthful populations. In general, mortality rates are low with strain B viruses. Less is known about the C strains, but they are not the cause of epidemics.
3.4 |
B Strain Viruses |
The first influenza B strain virus was isolated in 1940 during an epidemic in New York City. Influenza B virus seems to progress by causing localized outbreaks, chiefly in school children but sporadic cases do occur in adults, as well. It is very difficult to discern the level of antigenic variation in type B influenza. The viruses isolated during some of the larger outbreaks possessed variations within the haemagglutinin molecule. These variations were not comparable to any other previous forms of influenza B virus. This seems to suggest that once one form of B strain virus emerges, it is highly unlikely that another strain with similar serological qualities will return. The slow antigenic drift of the influenza B virus, unlike that of type A, allows time for human populations to acquire imminity, and eventually halt the spread of the virus.
3.5 |
Structure and Function of Selected Influenza Viral Proteins |
The neuraminidase antigen exists as a mushroom-shape spike on the surface of the virus. It has a box-like head consisting of four co-planar and roughly spherical subunits, and a hydrophobic region that is embedded within the interior of the viral membrane. Moreover, it is composed of a single polypeptide chain that is oriented in the opposite direction to the haemagglutinin antigen. The composition of the polypeptide is rather simple; a single chain of six conserved polar amino acids, followed by hydrophilic, nonconserved amino acids.
3.5.2 |
Other Viral Enzymes |
The P protein functions in several ways, all of which favor viral replication. It inhibits splicing of host cell M1 mRNA at the m3 5' site once it binds to the 5' end of the RNA. It permits binding of splicing factor 2 to purine-rich sequences in M1, and activates the sub-optimal M2 splice site.The PA, PB1, PB2, and NP proteins are transported to the host cell nucleus, first catalysing (+ ) strand RNAs, then (-) strand virion RNAs.
3.5.3 |
Influenza Virus Receptor |
Attachment of influenza A virus strains requires sialic acid. However, strains vary in their affinities for different sialyliogosacclarides. For example, avian virus strains prefer sialic acids attached to galactose with α-2,3 linkages while human virus strains prefer the α-2,6 linkages. Hemagglutinin (HA) is the viral glycoprotein that binds to the cell receptor, sialic acid. The HA monomer is synthesized as a precursor that is glycosylated and cleaved into HA1 and HA2 subunits. Each HA monomer consists of a long, helical chain anchored in the membrane by HA2 and topped by a large HA1 globule.
3.5.4 |
Replication and Spread of Infection |
Spread of infection is by direct contact. It is generally accepted that influenza viruses are maintained in humans by direct person-to-person contact. Infection begins in the upper respiratory tract by inhalation of droplets emanating from the respiratory tract of an infected individual. The virus first replicates in ciliated columnar epithelial cells of the respiratory tract. From there, they become included into respiratory secretions and spread by small-particle aerosols generated during sneezing, coughing, and speaking.
As soon as the virion encounters an epithelial cell of the respiratory tract, a receptor on the surface of that host cell interacts with the haemaggultinin on the surafce of the viral particle. The virion then attaches and is immediately included into a coated pit that develops within the cell membrane. The haemaggultinin then splits into two parts, H1 and H2, by a host cell protease. As the molecule is split, the coated pit containing the virion is transported into the cytoplasm. The viral core is then translocated into the nucleus where replication begins.
The incubation period for influenza (1-4 days) is short. The ability for the virus to infect large populations suggests that a single infected person can transmit the virus to a large number of susceptible individuals. Within a few hours after infection begins, 100 or more incomplete virions can be demonstrated by electron microscopy at the periphery of the cytoplasm just below the cell membrane. During that time, they are wrapped in a lipid coat, and eventually emerge as buds on the cell surface. They remain there until the neurominidase protein releases the mature viral particles into the interstitial space, where they can invade and destroy the next appropriate cell they encounter. Each invasion results in the death of the host cell, which is shed into the lumen of the respiratory tract.
3.6 |
Replication in Non-human Hosts |
Avian species provide a stable reservoir for maintaining viral gene sequences that emerge as recombinants for reintroduction into human populations. Avian influenza viruses have not changed much in antigenic structure in the last 60 years, in contrast to human and other mammalian flu strains that have undergone extensive antigenic variablity over that same time period. However, recent outbreaks in China among populations of domestic chickens with the rare H5/N1 strain illustrate the high degree of unpredicability regarding emergence of antigenic types with the avian flu viruses. In birds, influenza viruses replicate in the epithelial cells of the gastrointestinal tract, and are excreted in great quantity into the environment. Migrating waterfowl expose large numbers of domestic waterfowl to them each year on their way to their nesting grounds.
In humans, viral replication is restricted to the respiratory tract. Human strains of flu virus replicate in pigs, but not very well in birds. In contrast, the pig is a permissive host for both the human and bird strains. Because of this unusual feature, it is beleived that pigs are important in maintaining bird and human strains of the virus together in one place. Pigs are thought to serve as a “mixing vessel,” allowing for genetic exchange between avian and human viruses when the same cell is infected with both types of virus. Passage of human and avian strains of virus through pigs may also expose the virus to various selective pressures that allow antigenically unique, and often more virulent strains to emerge.
In China, pigs and waterfowl, such as domestic geese and ducks, are kept in close proximity to one another, and to large numbers of humans that tend them. This allows for the possibility of frequent infections in pigs with both human and bird strains of the virus. The results can be devastating. Nowhere else on earth is this situation present to such a degree. Hence, most epidemics of influenza are believed to have their origins in China.
3.7 |
Immune Responses During Infection |
Once the virus has penetrated the host cell, it initiates several types of viral-specific immune responses, including circulating and mucus membrane-associated secreted antibodies, interferon, and T-cell dependent responses. The latter can be divided into two types: T-helper cells and cytotoxic T-cells. Both antibody-based and cellular-based responses combine in neutralizing the neuraminidase and haemagglutinin portions of the virus. Interferon acts against viral RNA synthesis.
4 |
Ecology of Influenza: |
Tracking the epidemics and pandemics of the 19th century led the way for epidemiologists to devise a method of predicting distrubution patterns. Influenza epidemics and pandemics usually occur in one of four possible patterns: massive frontal movement, multiple sequester, seasonal epicenter relocation, and herald explosion.
4.1 |
Massive Frontal Movement |
This pattern consists of rapid diffusion of the disease from one or several areas during a single season, only. Diffusion within an area the size of Europe can occur in only a few months. Mortality rates associated with this kind of distribution pattern are usually high. In addition, future influenza events with the same strain are likely to occur, but they do not necessarily follow the same path. Some examples include the pandemics of 1580, 1732-37, 1782, and those of the early 1830’s.
4.2 |
Multiple Sequester |
This pattern also takes place within a given season and involves the formation of multiple epicenters along with an outward radial expansion of infection and disease. In contarst to massive frontal movement patterns, mortality from this variety of spread is coincident with low morbidity rates and lower rates of diffusion. The pandemic of 1847-48 is a good example of the multiple sequester type of outbreak.
4.3 |
Seasonal Epicenter Relocation |
This pattern does not necessarily involve pandemics, but it is characterized by a relocation of epicenters from one geographic region to another depending on the season. The European outbreak of 1803 best fits this pattern.
4.4 |
Herald Explosion |
This pattern of emergence consists of a combination of all three patterns, and includes a seasonal spring wave, presumed to initiate in remote locations. While initial mortality rates appear low during spring, herald explosion patterns travel large distances within a few months. A second wave may ensue during autumn or winter, with increased mortality rates. The pandemic of 1842-43 typifies this pattern.
5 |
Treatment and Prevention Of Influenza Outbreaks |
The guiding principal of any antiviral drug development is to inhibit viral replication, thus dramatically reducing the number of infectious particles. Antiviral drugs have been available since the early 1980’s, but they are not very effectiveness against the influenza virus, due to its rapid development of resistance to them. Thus, current treatments for influenza are limited.
An inactivated influenza vaccine is currently available and is the most widely used method to prevent influenza outbreaks in high-risk populations, such as the elderly. Vaccines elicit immune responses that attack the neuraminidase and haemagglutinin proteins found on the surface of the influenza virus. Anti-neuraminidase antibodies decrease the severity of disease, while anti-haemaggultinin antibodies neutralize viral infectivity.Yet, vaccination only provides protection against outbreaks involving known viral strains. As mentioned, because of its penchant to vary its antigenic components from year to year, vacination can prove ineffective.
5.1 |
New Antiviral Drugs |
Adamantamine was the first highly specific, potent antiviral drug effective against any virus. It specifically inhibits influenza A virus. The target of the drug is the H2 protein, a tetrameric transmembrane ion channel that transports proteins. The drug blocks the channel so that proteins cannot enter the virus. Adamantamine is highly specific for type A influenza but not type B because the latter does not contain an M2 protein. The active site of neuraminidase is essential for viral replication, and thus highly conserved. This allows for antiviral drug design, targeting the active site without major concern regarding the development of resistance.
One chemical agent currently being tested is a carbocyclic compound (3,4R,5S)-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexane-1-carboxylic acid, also known as Ro6408802. This compound is worth mentioning because it fits into the three-dimensional structure of the neuraminidase active site and completely inhibits the enzyme. Although these two drugs are not currently available, they are now in preclinical and clinical drug trials, and give encouragement towrds developing an effective chemotherapeutic approach to the treatment of influenza.
5.2 |
Predicting Emergence of Influenza Outbreaks |
As millions died from the 1918-1920 influenza pandemic, there was a growing need to recognize the possibility for future influenza outbreaks more quickly in the future. One major surveillance method was championed by Robert Serfling, chief of statistics in the Epidemiology Bureau of the Communicable Disease Center. According to Serfling, three things were required to determine “advance estimation of expected pneumonia-influenza mortality levels”: 1) determination of a “secular trend”, 2) an estimation of seasonal variations and 3) a distinction between major influenza cycles and random variations.
Serfling’s Model is a curvilinear graph which utilizes a sine/cosine in the estimation of an influenza epidemic occurring for a given influenza season. One of the first successful applications of this method was to accurately estimate the magnitude of the 1957 Asian influenza pandemic. This regression technique is still widely practiced throughout the United States and has been employed in other countries, as well.
5.3 |
Serfling's Model |
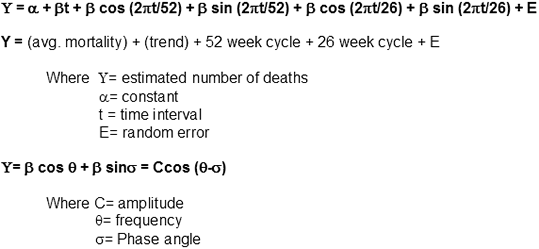
6 |
Avian Influenza A |
Introduction
Influenza is a common, usually mild infection in non-domestic waterfowl that can cause serious disease in domestic waterfowl and humans. Myriad strains are ubiquitous in animal reservoirs, hence its ecology encompasses all those wildlife networks favouring the maintenance of strain diversity among wild bird populations. Unlike other highly contagious viruses, such as measles and smallpox, influenza viruses have the potential for continuous antigenic shift and drift due to their unique segmented viral genomes. These general biological characteristics help set the stage for its potential for causing pandemic outbreaks in humans, as well as in avian populations. Understanding the interfaces between host susceptibility, strain diversity, and animal reservoirs is critical for developing models for prediction of epidemics, and their control and prevention.
The Virus: Classification of Influenza A Viruses
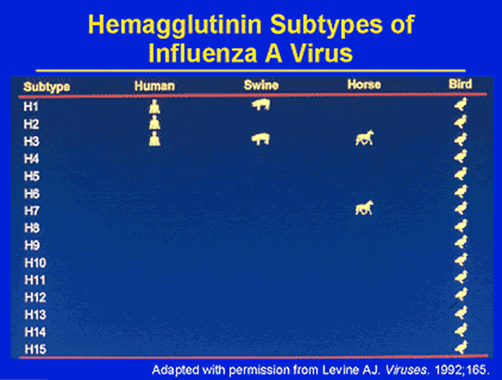
This classification system has characterized pandemics of the 20 th century: H1N1 Spanish flu in 1918, H2N2 Asian flu in 1957, H3N2 Hong Kong flu in 1968 and an H1N1 Russian flu in 1977 with the ethnicities attached as somewhat misnomers, indicating only the location in which epidemic case numbers were first documented. One hypothesis suggested that pandemics could be caused by a viral strain to which humans were naïve (i.e., had no immunity), and that these new antigenic types arose from antigenic drift of human viruses and/or genetic recombination of human viruses with animal viruses in a “mixing vessel”, usually pigs.
Why are swine ‘mixing vessels'?
HA binds to various sialic acids depending upon species specificity and the linkages of these sialic acids to galactose on host surface cells. While avian influenza viruses bind preferentially to one type of linkage with sialic acid (alpha 2,3) human viruses bind primarily to another (alpha 2,6), and swine influenza viruses can bind to either of these linkages, creating a ‘mixing vessel' for viruses of birds and humans. Essentially, pigs are mixing vessels because their respiratory epithelial cells have the receptors for both human and avian influenza viruses, which allows for co-infection and subsequent recombination events. However, humans could very well become ‘mixing vessels' since the airway epithelial cells in humans contain receptors for both avian and human influenza viruses.

What determines virulence of influenza viruses?
Although strain types are characterized by their H and N surface glycoproteins, strain pathogenicity is also dictated by other factors, including the gamut of H, N, viral genes, and host factors (e.g., immune response genes, age, sex), as well.
 |
Influenza strain diversity is defined by two glycoproteins on the surface of the virion. The first stage of infection with the virus is mediated by the hemagglutinin (HA) membrane glycoprotein that allows for binding and fusion with host cells. The other major glycoprotein, neuraminidase, facilitates viral budding and exit from the host cell. These two glycoproteins exhibit great structural diversity, and at least 15 hemagglutinin and 9 neuraminidase subtypes have been identified to date. However, only viruses of hemagglutinin subtypes H1, H2, and H3 and neuraminidase subtypes of N1 and N2 are currently circulating in the human population. Avian species are considered the predominant natural hosts for viruses composed of other glycoprotein subtypes (see diagram) although a variety of animal species have been infected with various viruses.
This theory was challenged in 1997 when an avian influenza strain H5N1 infected 18 people in Hong Kong, with a 33% case-fatality rate. This highly virulent strain of chicken influenza, H5N1, was of avian origin and had not re-assorted with human viruses, infecting humans in direct contact with those domestic fowl. Since 1997, several mini-outbreaks of H5N1 have occurred each year in Southeast Asia, with similar mortality rates. Fortunately, the avian origins of this virus have not allowed for person-to-person transmission, although case reports in early 2005 suggested the possibility of person-to-person transmission in rural China.
The outbreak as of August, 2005
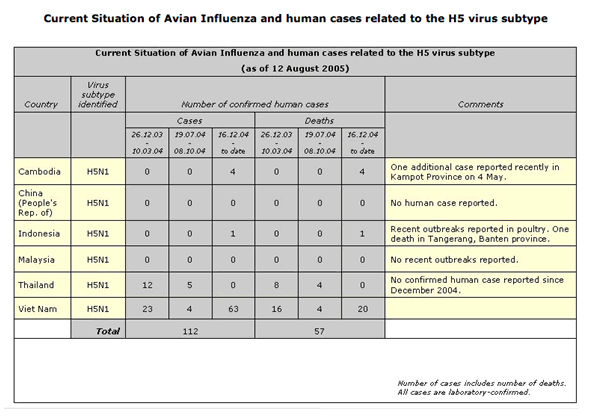

Avian viral strain H9N2 in 1998 also caused human infection. This virus bore no evidence of recombination, even though its binding properties more closely resembled that of human influenza virus origin rather than from any avian strain. Illness was “mild and self-limited”. An avian virus, H7N7, in the Netherlands in 2003, caused an outbreak amongst poultry farmers and their families. Although this strain was not particularly pathogenic, person-to-person transmission was documented. Taking this data into account, in the late 1990s, a shift in thinking about the nature of influenza outbreaks allowed for the possibility that avian viruses are indeed potential pandemic threats.
What should we do?
How can the scientific community predict which strain is the most likely candidate to cause the next pandemic? Equally important, how can the world best prepare for such an event? The H1N1 and H3N2 are the current primary circulating strains in the human population, but the high virulence of the H5N1 strain has elicited speculation as to whether this or other related avian strains are capable of causing pandemic influenza. In order for any of the avian influenza viruses to evolve into one capable of person-to-person transmission, many changes in the genome of the virion would have to precede such an event. The current person-to-person capability of the virulent H5N1 is extremely low or non-existent, due to the fact that the hemagglutinin molecule is not configured properly to interact with human cell surface receptor proteins of the respiratory tract. The virulence of the transmissible H7N7 is mild, so the probability is low of these strains becoming a major worldwide health risk. Noteworthy is the fact that humans do not have any immunity to H5, hence this strain is highly pathogenic, and in many cases lethal. Even though the odds are small of the H5N1 strain becoming capable of human to human transmission, the accumulation of a series of point mutations could eventually allow for person-to-person spread, and could thus cause a pandemic even greater than 1918. Whether ‘mixing' vessels, antigenic drift, or other mutational accumulations will lead to its emergence cannot yet be determined. In addition, the continuing circulation of H1N1 and H3N2 raises further concerns as to their epidemiological fate.
Vaccines
Influenza vaccine development is an unreliable variable in helping to quell viral pandemicity. Annual worldwide vaccine usage of over 300 million doses requires more than six months and 350 million chicken eggs to meet that demand. These constraints and other extraneous factors, such as the microbial contamination of vaccines manufactured in the past year in the UK , leaves influenza vaccine technology currently in a state of disconnect with imminent pandemic threats and the corresponding health demands. The detection of a novel strain in the human population may render subsequent vaccine development too long a process on which to rely for protection. Current vaccines (killed-virus) contain H1 and H3 components. Obviously, the lack of other HA components in these vaccines is a major flaw in the control strategy, particularly with respect to the number of recent avian influenza outbreaks in humans. Alternatively, since H2 has also established a propensity for infection in humans, this strain should be considered as part and parcel of the standard antigenic mix.
Although these pandemic flu scenarios depict a less than satisfactory situation, several measures in preparation for such an event may mitigate or prevent such a pandemic. Continued surveillance efforts both in humans (particularly in Southeast Asia ) and animal (avian) species must be stepped up and reinforced. Continued research into vaccine design, development, and rapid scale-up production methods is essential. Other efforts could include the stockpiling of antiviral drugs, and a greater commitment by governments to an early warning system.
Why is the current influenza vaccine technology unreliable?
Influenza virus is a ‘shape-shifter,' so every year new strategies must be devised to design a vaccine against the predicted flu strain for that year. This has proven difficult because of the uncertainty of which influenza strain will actually emerge. Frequently, these vaccines are based on the previous year's strains. Ultimately, researchers are trying to design a vaccine that could confer immunity to all influenza strains. However, as of yet, this ‘Holy Grail' has not been discovered. The prospects for a DNA-based vaccine is also being investigated, as well as RNA interference, and new antivirals.
Summary of the ecology of influenza viruse

Selected References
Gamblin, S.J. et al. 2004. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science, 303:1838-1842.
Kaiser, J. 2004. Searching for all-powerful flu weapons. Science , 306:395.
Koopmans, M. et al. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands . Lancet, 363: 587-593.
Laver, G., Garman, E. 2001. The origin and control of pandemic influenza Science, 293: 1776-1777
Liem, N.T. et al. 2005. Lack of H5N1 avian influenza transmission to hospital employees, Hanoi , 2004. Emerging Infectious Disease ,11: 2,210-215.
Nicholson, KG et al. 2003. Influenza. Lancet , 362:1733-1745.
Osterholm, M.T. 2005. Preparing for the next pandemic. New England Journal of Medicine 352: 1839-1842
Palese, P. 2004. Influenza: old and new threats. Nature Medicine Suppl. 10:12, S82-S87.
Suzuki, Y. 2005. Sialobiology of influenza L molecular mechanism of host range variation of influenza viruses. Biol Pharm Bull , 28:399-408.
Ungchusak, K. et al. 2005. Probable person-to-person transmission of avian Influenza A (H5N1) New England Journal of Medicine, 352:4,333-340.
Web Sites of Interest
http://europa.eu.int/comm/health/ph_threats/com/Influenza/avian_influenza_en.htm
http://www.promedmail.org/pls/promed/f?p=2400:1600:15323012213163428114
http://gamapserver.who.int/GlobalAtlas/home.asp
http://www.who.int/csr/disease/avian_influenza/en/